About UniPlant-CG
Why UniPlant-CG?
Accurately classifying and grading plant diseases in real-world settings is challenging due to domain shifts, unseen species, and limited labeled data. As illustrated in Fig. 1, Uniplant-CG addresses these issues through a unified multi-task framework that jointly learns species, disease, and severity representations. By combining prototype-guided learning with cross-domain feature alignment, it adapts to new environments and novel symptoms without requiring target-domain labels, thereby enhancing the robustness and practicality of plant disease diagnosis and management.
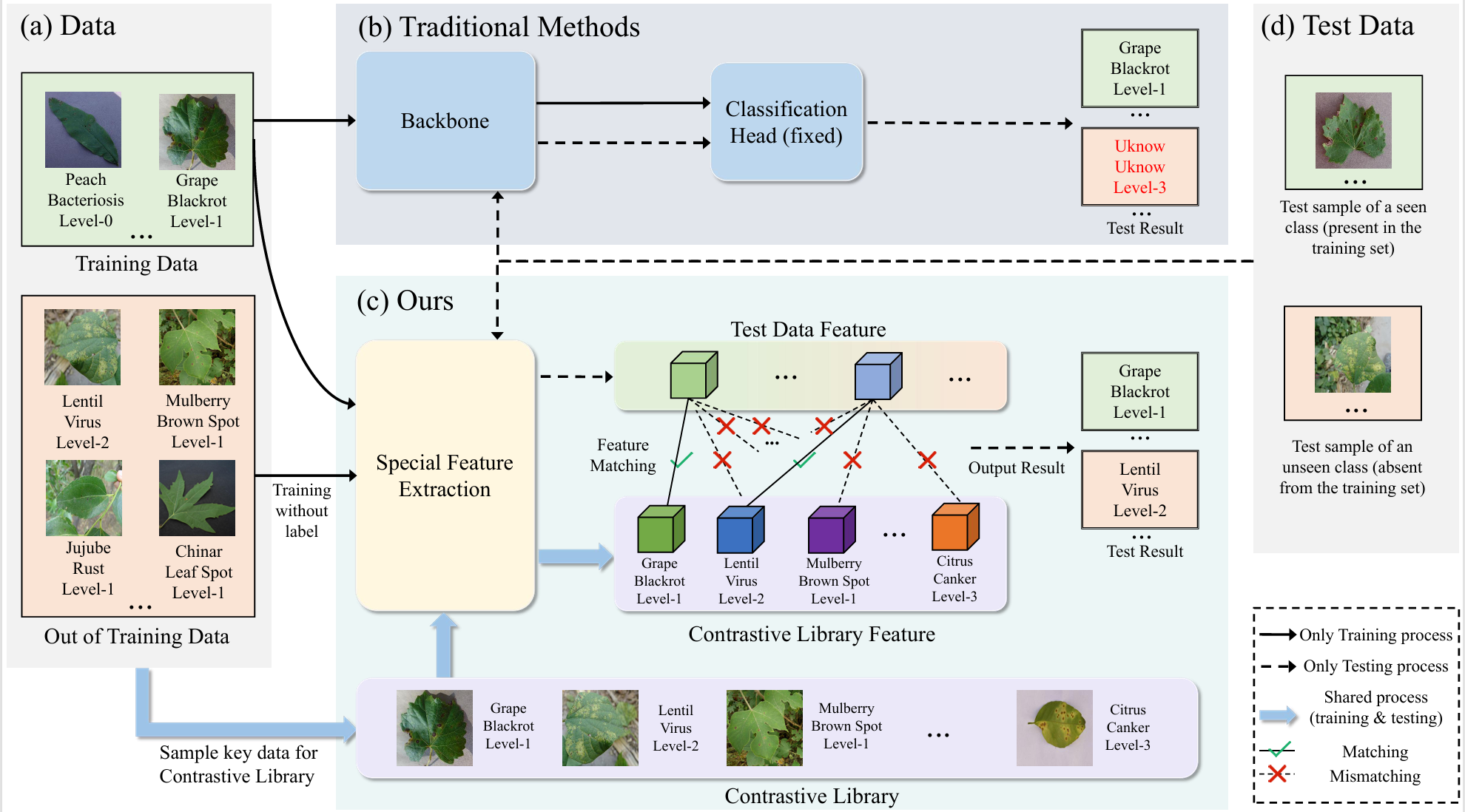
Figure 1: Comparison of open-set plant disease recognition between Uniplant-CG and traditional methods. Unlike traditional models that struggle with unseen species and domain shifts, Uniplant-CG effectively generalizes to new conditions through prototype-guided learning and cross-domain alignment, achieving more stable and reliable recognition in open-world scenarios..
Application and Prospects
Unified frameworks like Uniplant-CG advance intelligent plant disease management by enabling simultaneous species identification, disease recognition, and severity grading under real-world, open-set conditions. Through prototype-guided learning and cross-domain adaptation, the model provides reliable diagnosis across diverse environments without requiring additional labeling. This capability supports precision agriculture by facilitating accurate, scalable, and generalizable plant health assessment. Future work will focus on integrating Uniplant-CG into real-time monitoring systems and expanding its adaptability to broader crop varieties and dynamic field conditions, further enhancing its role in sustainable agricultural development..
Conclusion
We propose a unified framework for plant disease recognition and severity estimation that generalizes to unseen domains. Leveraging a dual-branch contrastive design, the model captures both global context and localized lesion patterns. Its modular structure allows flexible adaptation to species classification, disease detection, and severity grading by updating the contrastive feature library and task-specific parameters, without altering the overall architecture. Experiments on a large-scale annotated dataset demonstrate strong cross-domain performance and state-of-the-art accuracy. This approach offers a practical solution for automated plant health monitoring and provides a foundation for future work on open-set recognition, rare category adaptation, and interpretable AI-based diagnosis.
Research Team
SAMLab
