Uniplant-CG: Knowledge-Driven Unified Framework for Plant Disease Classification and Severity Grading Real farms present an open-world setting with shifting sensors, sites, and seasons, making disease recognition and grading hard to generalize. Our system addresses this by pairing a contrastive library that captures cues for never-seen symptoms with a cross-domain alignment module that stabilizes features across data sources. Built on Leaf-CG—about 441,488 expert-labeled images covering 59 plant species, 373 disease types, and four severity levels—the model handles species ID, disease recognition, and severity estimation in one pipeline. In benchmark studies it surpasses recent baselines on in-domain and out-of-domain tests (e.g., higher overall accuracy and a markedly stronger true-positive rate for open-world detection), without using target-domain labels, pointing to practical readiness for precision agriculture.
overview: We propose a unified framework for plant leaf disease classification and severity grading under open-set conditions, as shown in Fig. 1. It combines multi-level contrastive learning for labeled data, enabling the model to learn basic knowledge of plant species, diseases, and severity. A contrastive feature library anchors novel class identification, while a cross-domain adapter uses unlabeled out-of-domain data to align feature distributions. By expanding the label space without retraining the entire model, our framework provides robust generalization to unseen categories, improving plant disease diagnosis across diverse conditions.
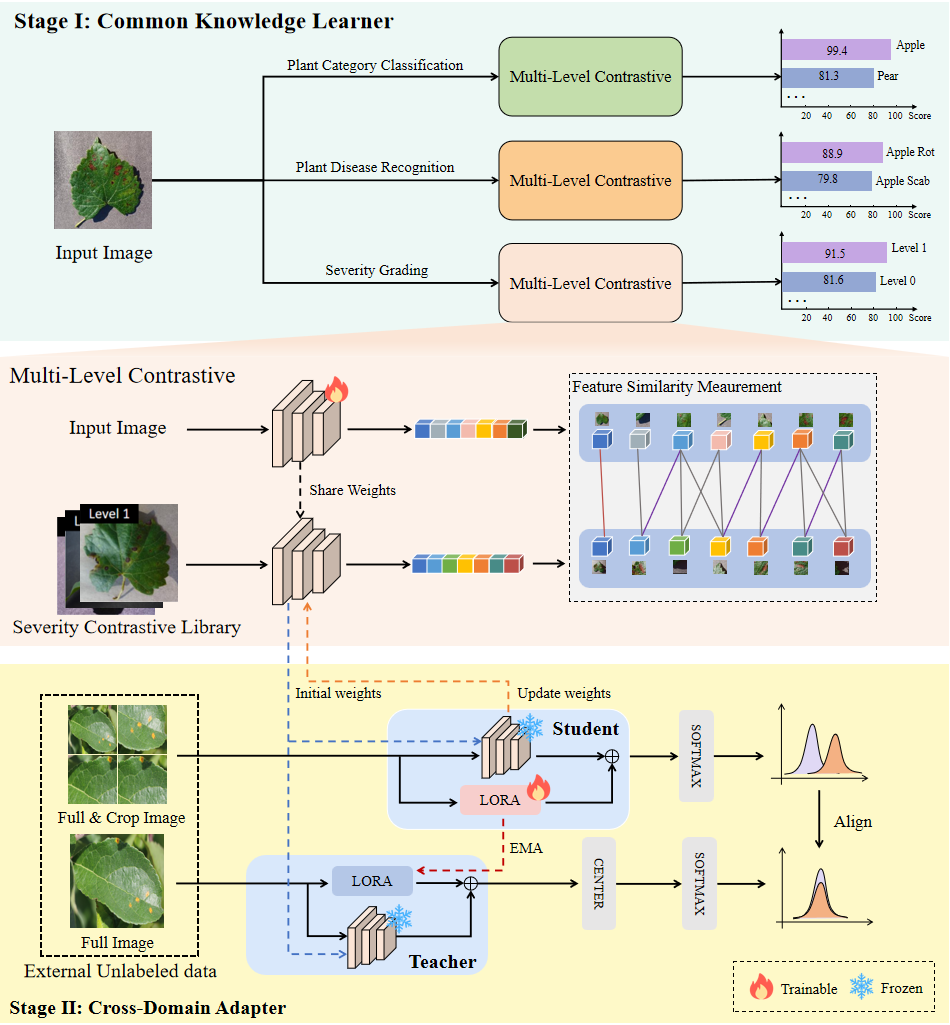
Figure 1: The Uniplant-CG framework. Including Common Knowledge Learner Module, Multi-Level Contrastive Module and Cross-Domain Adapter Module.
Common Knowledge Learner Module The Common Knowledge Learner (CKL) is the first module in our framework, designed to help the model recognize plant species, disease types, and severity levels. It uses contrastive learning to match input features with a curated set of reference embeddings, improving generalization to new or rare categories. This stage forms the core foundation for more advanced tasks like domain adaptation.
Multi-Level Contrastive Module The Multi-Level Contrast (MLC) component augments CKL by comparing features across scales—from whole-image semantics down to localized lesion details—against a library of representative embeddings. This hierarchical matching enriches the representation space, making the model better at distinguishing both frequent and scarce disease types and more sensitive to the subtle visual cues needed for accurate severity estimation.
Cross-Domain Adapter Module We design the Cross-Domain Adapter (CDA) to help the model adapt to unseen data distributions. Built on a lightweight student–teacher strategy, CDA leverages unlabeled samples to refine feature alignment across domains. This process allows the model to transfer knowledge from familiar conditions to new environments while maintaining the recognition performance established during the CKL stage.
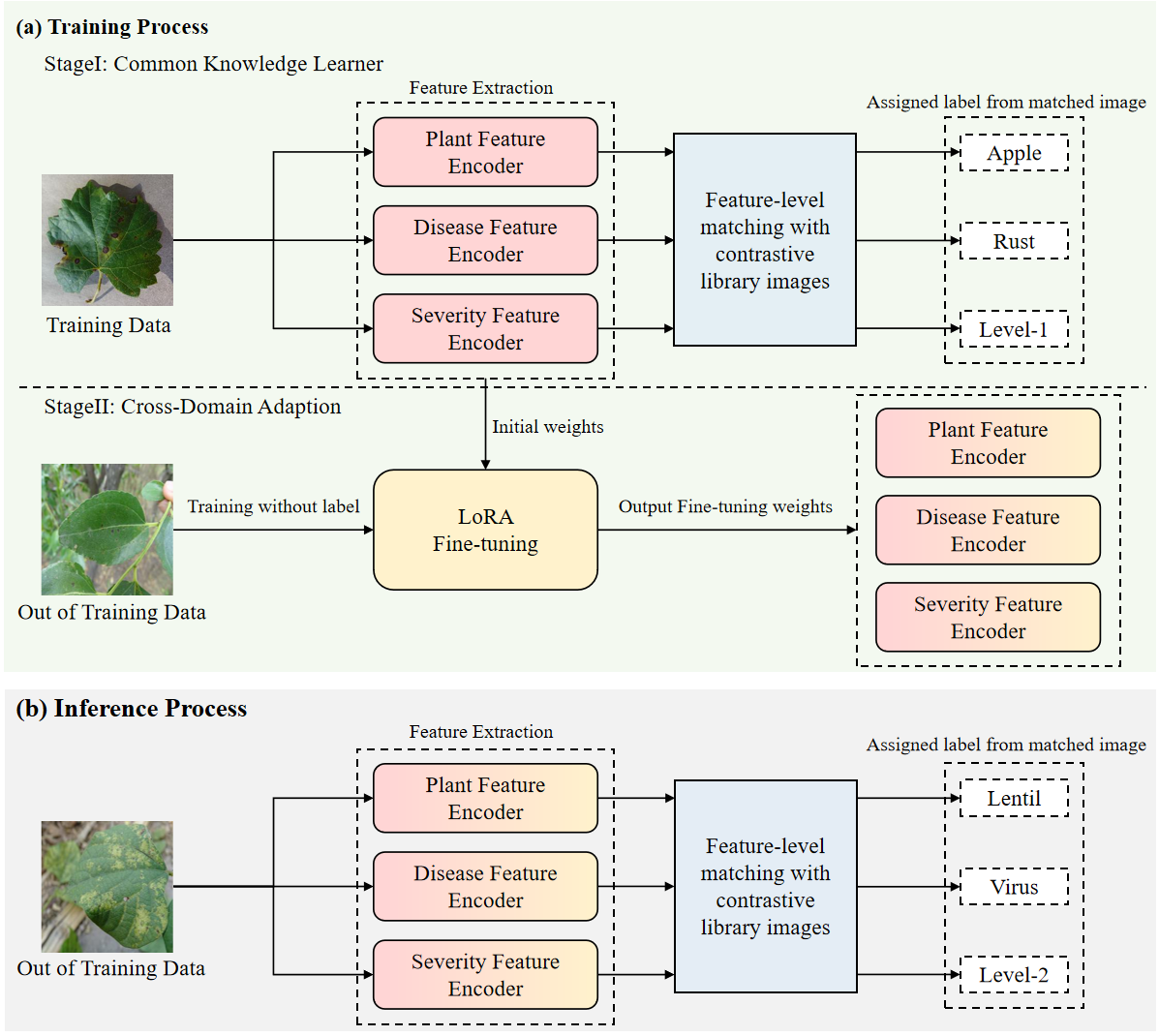
Figure 2: The inference process diagram of our method.
